Research Article
Magdalena Jędrzejczak-Silicka , Krystyna Dudaniec, Andrzej Dybus
Laboratory of Cytogenetics, West Pomeranian University of Technology, Szczecin, Klemensa Janickiego 29, 71-270 Szczecin, Poland
Abstract. The objective of the study was to investigate possible effect of the polymorphism in pigeon αA-globin (AGLOB) gene on its expression level. The g.5768C>T substitution located in 3′ flanking region of the gene was analyzed. PCR-RFLP (AvaII) method was used to identify the SNP. The gene expression analyses were performed using qPCR. The investigation indicated a difference but not significant in the αA-globin gene expression levels between pigeons carrying different g.5768C>T genotypes. The AGLOBCC genotype was associated with 2.46-fold higher expression level of pigeon αA-globin gene compared to AGLOBTT genotype. The lower expression of αA-globin gene in AGLOBTT pigeons may be caused by the presence of cis-acting regulatory elements in the analysed gene region.
Keywords: αA-globin gene, racing pigeons, qPCR, SNP
Pigeons have developed a strong relation with humans. There is evidence that cave-dwelling humans exploited Rock Doves for food at least 67 thousand years ago [Blasco et al. 2014Blasco, R., Finlayson, C., Rosell, J., Marco, A.S., Finlayson, S., Finlayson, G., Negro, J.J., Pacheco, F.G., Vidal, J.R. (2014). The earliest pigeon fanciers. Sci. Rep., 4, 5971. https://doi.org/10.1038/srep05971.]. In the ancient Middle East, doves were kept in towers; their droppings with high nitrogen content were a major source of organic fertilizer [El Gemaiey 2016El Gemaiey, G.A.M. (2016). The Pigeon Towers of Isfahan-Iran. IOSR-JHSS., 21(12), 69–81.]. People used pigeons to send war reports home from the front. The discovery of the pigeon’s homing ability opened unforeseen possibilities that extend beyond that of a messenger. Nevertheless, sports and gambling are as old as war [Jerolmack 2007Jerolmack, C. (2007). Animal archeology: Domestic pigeons and the nature-culture dialectic. Qual. Sociol. Rev., 3(1), 74–95.], therefore intensive management and breeding for meat or sport, isolate pigeons from external selective pressures were performed [Marom et al. 2018Marom, N., Rosen, B., Tepper, Y., Bar-Oz, G. (2018). Pigeons at the edge of the empire: Bioarchaeological evidences for extensive management of pigeons in a Byzantine desert settlement in the southern Levant. PLoS One, 13(3), e0193206. https://doi.org/10.1371/journal.pone.0193206.]. Moreover, sport pigeons are under continuous selection to improve their speed, spatial orientation, and endurance during long flights, however, there are many factors affect these traits in racing pigeons [Ramadan et al. 2018Ramadan, S., Miyake, T., Yamaura, J., Inoue-Murayama, M. (2018). LDHA gene is associated with pigeon survivability during racing competitions. PLoS One, 13(5), e0195121. https://doi.org/10.1371/journal.pone.0195121.], so searching a reliable and precisely gene marker is crucial for now.
There are many tools for selection, but the most common types of markers in genomes are SNPs [Goldstein and Cavalleri 2005Goldstein, D.B., Cavalleri, G.L. (2005). Genomics: Understanding human diversity. Nature, 437(7063), 1241–1242. https://doi.org/10.1038/4371241a., Jacob et al. 2018Jacob, Y., Spiteri, T., Hart, N.H., Anderton, R.S. (2018). The potential role of genetic markers in talent identification and athlete assessment in elite sport. Sports (Basel), 6(3), 88. https://doi.org/10.3390/sports6030088.], nowadays, genetic markers have been used to predict racing performance in pigeons [Proskura et al. 2014Proskura, W.S., Cichoń, D., Grzesiak, W., Zaborski, D., Sell-Kubiak, E., Cheng, Y-H, Dybus, A. (2014). Single nucleotide polymorphism in the LDHA gene as a potential marker for the racing performance of pigeons. J. Poult. Sci., 51(4), 364–368. https://doi.org/10.2141/jpsa.0130237., Proskura et al. 2015Proskura, W.S., Kustosz, J., Dybus, A,. Lanckriet, R. (2015). Polymorphism in dopamine receptor D4 gene is associated with pigeon racing performance. Anim. Genet., 46(5) 586-587. https://doi.org/10.1111/age.12328., Proskura et al. 2017Proskura, W.S., Łukaszewicz, A., Dzierzba, E., Cichoń, D., Zaborski, D., Grzesiak, W., Dybus, A. (2017). The Cys83Gly amino acid substitution in feather keratin is associated with pigeon performance in long-distance races. Vet. Med., 62(4), 221–225. https://doi.org/10.17221/271/2015-vetmed., Ramadan et al. 2018Ramadan, S., Miyake, T., Yamaura, J., Inoue-Murayama, M. (2018). LDHA gene is associated with pigeon survivability during racing competitions. PLoS One, 13(5), e0195121. https://doi.org/10.1371/journal.pone.0195121., Dybus et al. 2018Dybus, A., Proskura, W., Pawlina, E., Nowak, B. (2018). Associations between polymorphisms in the myostatin, αA-globin and lactate dehydrogenase B genes and racing performance in homing pigeons. Vet. Med., 63(8), 390–394. https://doi.org/10.17221/149/2017-vetmed.]. Recently, in whole-genome study, the CASK gene was marked as an important genetic factor for fast flight, long endurance, and accurate navigation was revealed [Gazda et al. 2018Gazda, M.A, Andrade, P., Afonso, S., Dilyte, J., Archer, J.P., Lopes, R.J., Faria, R., Carneiro, M. (2018). Signatures of selection on standing genetic variation underlie athletic and navigational performance in racing pigeons. Mol. Biol. Evol., 35 (5), 1176–1189. https://doi.org/10.1093/molbev/msy030.].
There are a large number of polymorphisms/mutations in globin gene in hemoglobinopathies context [Alaithan et al. 2018Alaithan, M.A., Abdul Azeez, S., Borgio, J.F. (2018). A comprehensive review of the prevalence of beta globin gene variations and the co-inheritance of related gene variants in Saudi Arabians with beta-thalassemia. Saudi Med. J., 39(4), 329–335. https://doi.org/10.15537/smj.2018.4.21360.]. In birds, Galen et al. [2015Galen, S.C., Natarajan, C., Moriyama, H., Weber, R.E., Fago, A., Benham, P.M., Chavez, A.N., Cheviron, Z.A., Storz, J.F., Witt, C.C. (2015). Contribution of a mutational hot spot to hemoglobin adaptation in high-altitude Andean house wrens. Proc. Natl. Acad. Sci. USA, 112(45), 13958–13963. https://doi.org/10.1073/pnas.1507300112.] reported that a single point mutation in the βA-globin gene changed the hemoglobin function. Targa et al. [1993Targa, F.R., de Moura Gallo, C.V., Huesca, M., Scherrer, K., Marcaud, L. (1993). Silencer and enhancer elements located at the 3′-side of the chicken and duck alpha-globin-encoding gene domains. Gene, 129(2), 229–237. https://doi.org/10.1016/0378-1119(93)90273-6.] identified regulatory elements at the 3′-side of the chicken and duck α-globin gene domains. Furthermore, García-González and Recillas-Targa [2014García-González, E., Recillas-Targa, F. (2014). A regulatory element affects the activity and chromatin structure of the chicken α-globin 3′ enhancer. Biochim. Biophys. Acta., 1839(11), 1233–1241. https://doi.org/10.1016/j.bbagrm.2014.09.009.] described regulatory element which modulates the activity of the chicken α-globin 3′ enhancer. In pigeons, two SNPs were detected in αA-globin gene, one of them in 3′ region of the gene [Dybus et al. 2008Dybus, A., Ming-Huang, Ch., Cheng, Y-H., Szatkowska, I. (2008). DNA Polymorphism of the α-A globin Gene in Domestic Pigeon. Anim. Sci. Pap. Rep., 26(3), 3219–322.].
The aim of this study was to investigate the association between polymorphism in 3′ region of the αA-globin gene and relative expression level of the gene in the three genotype group of racing pigeons.
Ten homing pigeons (5 hens and 5 cocks) averaged six months old from the West Pomeranian University’s loft were used in the study. The pigeons used in this study were reared in one loft and feed upon the same diet of dry mixed grains (35 grams per pigeon a day), free access to minerals (grit) and water. For the expression gene analysis ten pigeons were selected from the group of all individuals (~70) kept in the loft, by previous genotyping [Dybus et al. 2008Dybus, A., Ming-Huang, Ch., Cheng, Y-H., Szatkowska, I. (2008). DNA Polymorphism of the α-A globin Gene in Domestic Pigeon. Anim. Sci. Pap. Rep., 26(3), 3219–322.]. Gender identification was determined as described by Griffiths et al. [1998Griffiths, R., Double, M.C., Orr, K., Dawson, R.J.A. (1998). DNA test to sex most birds. Mol. Ecol., 7(8), 1071–1075. https://doi.org/10.1046/j.1365-294x.1998.00389.x.]. The analysed polymorphic site is located in the 3′ flanking sequence (at a distance of 65 nucleotides from the end of the 3′UTR) of the αA-globin gene [NW_004973488.1].
DNA was isolated from whole blood using salting-out procedure (Epicentre Biotechnologies, Madison, WI, USA). The PCR-RFLP method (with AvaII enzyme) was used to identify the pigeons’ g.5768C>T SNP in the 3′ region of the αA-globin gene [NW_004973488.1]. PCRs were carried out in 15 µl (~60 ng of DNA, 15 pmol of each primer, 1.5 µl of 10×PCR buffer, 1.5 mM MgCl2, 200 µM dNTP, 0.3 unit of Taq polymerase; EURX, Gdańsk, Poland). The following thermal profile was applied: 5 min / 94°C followed by 33 cycles (94°C / 20 s, 61°C / 30 s and 72°C / 30 s) and 72°C / 5 minutes. Amplicons were digested with 2 units of AvaII (New England Biolabs, Beverly, MA, USA) at 37°C and separated (120 V, 50 min) in 4% agarose gel (Sigma-Aldrich, St Louis, MO, USA).
Total RNA was extracted from the 3 µl of blood immediately after its collection (Total RNA Kit, A&A Biotechnology, Poland). RNA samples were digested with 1 unit of DNase I (Qiagen, Hilden, Germany) to remove possible DNA contamination. Total RNA was finally eluted in 100 µl of RNase-free dH2O (AppliChem, Darmstadt, Germany). RNA concentration was assessed (mean of three measurements) with the Qubit 2.0 fluorometer (Quant-iTTM RNA BR Assay Kit; Invitrogen, Carlsbad, CA, USA). Two-step reverse transcription was performed using TranScriba Kit based on Oligo-dT18 primers and MMLV reverse transcriptase (A&A Biotechnology, Poland) according to the vendor’s protocol. Synthesized cDNA was stored in –20°C.
Intron-spanning primers bind in the exon 1 and 2 and led to amplify a 131 bp fragment of the AGLOB cDNA. The qPCR analyses were performed in Rotor Gene 6000 thermocycler (Corbett Research, UK). The qPCR mixture contained 7.5 µl Kapa Sybr Fast qPCR Mix (KAPA Biosystems, USA), ~10 ng of cDNA, 200 nM of each primer and nuclease-free water (AppliChem, Germany) up to 15 µl volume. The qPCR conditions were: 3 min / 95°C followed by 40 cycles (95°C / 3 s, 60°C / 3 s, 72°C / 20 s) and 72°C for 3 minutes. All samples were run in triplicates. No template control was included in every qPCR run.
The differences in relative level of AGLOB expression between the genotypes were tested using Kruskal Wallis non-parametric ANOVA, while the differences between sexes were analysed using Mann-Whitney U test [Livak and Schmittgen 2001Livak, K.J., Schmittgen, T.D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods, 25(4), 402–408. https://doi.org/10.1006/meth.2001.1262., Pfaffl 2001Pfaffl, M.W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res., 29(9), e45. https://doi.org/10.1093/nar/29.9.e45.]. All PCR primers (for PCR-RFLP and qPCR) were designed with Primer3 software [Untergrasser et al. 2012Untergrasser, A., Cutcutache, I., Koressaar, T., Ye, J., Faircloth, B.C., Remm, M., Rozen, S.G. (2012). Primer3 – new capabilities and interfaces. Nucleic Acids Res., 40(15), e115. https://doi.org/10.1093/nar/gks596.] and presented in Table 1.
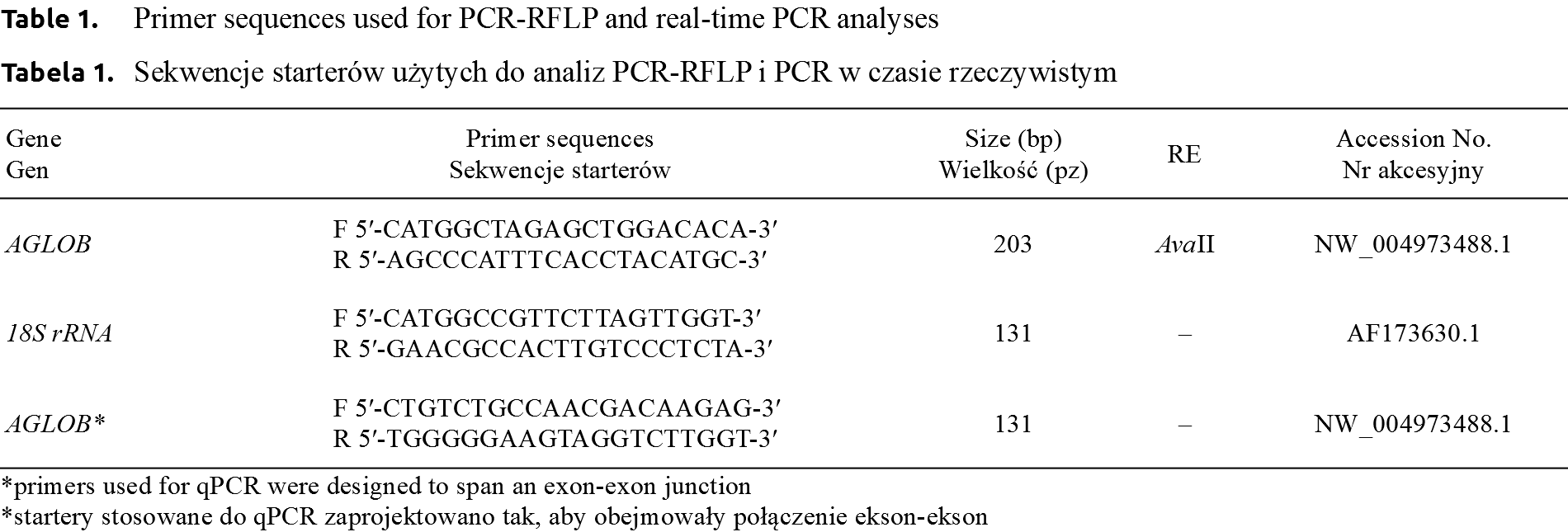
The PCR-RFLP was performed to identify the genotypes (AGLOBTT – 165 and 38 bp, AGLOBCT – 165, 152, 38, 13 bp, AGLOBCC – 152, 38, 13 bp) in all young pigeons kept at the University’s loft. Sex of pigeons was determined by PCR using CHD1 primers (Fig. 1) [Griffiths et al. 1998Griffiths, R., Double, M.C., Orr, K., Dawson, R.J.A. (1998). DNA test to sex most birds. Mol. Ecol., 7(8), 1071–1075. https://doi.org/10.1046/j.1365-294x.1998.00389.x.]. However, only ten individuals with no relationship based on the accurate pedigree were selected for qPCR analysis. The relative expression level of AGLOB gene was determined in selected six-month individuals (5 from both genders). The average AGLOB gene expression level, which is calculated by the average relative quantity (RQ of AGLOB gene expression was normalized using 18S rRNA gene expression values) in the females group was 1.58-fold higher than that of males, the average Ct values were 19.05 and 20.87 for females and males groups, respectively. Only numerical, not statistical significance differences between gender were confirmed.
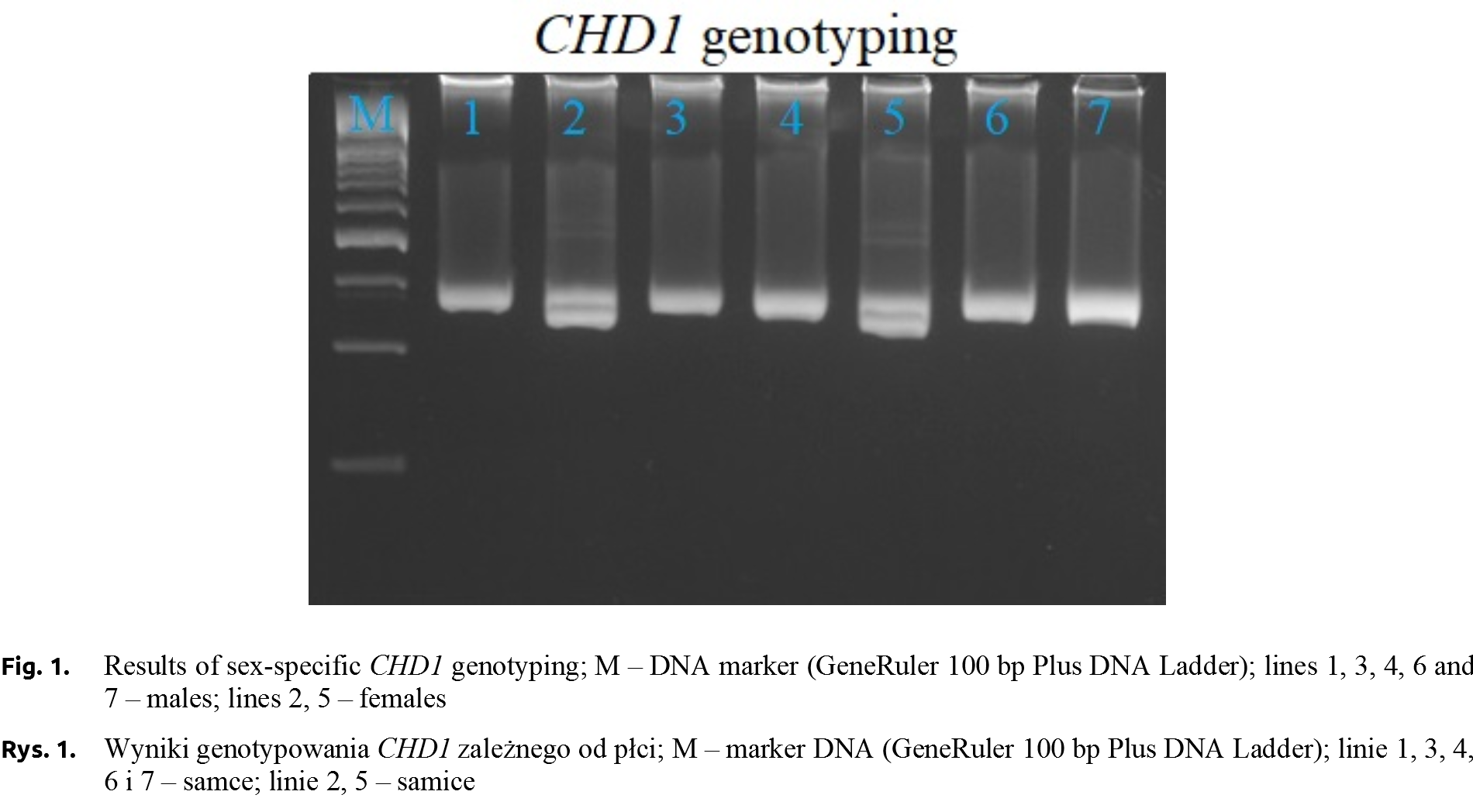
RQ of AGLOB gene expression analysis focused on individuals divided into three genotype groups. The average Ct values in group of AGLOBTT and AGLOBCT individuals were slightly similar (20.76 and 20.45, respectively). The lowest values (18.99) were observed for AGLOBCC ones. The relative expression level of AGLOB gene (normalized using 18S rRNA gene expression values) was the highest in the AGLOBCC individuals (RQ = 0.65). The RQ value for AGLOBCT pigeons was 0.57 and the lowest was recorded in the AGLOBTT (RQ = 0.26, Fig. 2).
Gene expression can be controlled at the transcriptional and translational level through the characteristic non-coding DNA sequences called cis-regulatory elements. These elements may be located in the vicinity or at a long distance from the coding sequence [Daniel et al. 2014Daniel, B., Nagy, G., Nagy L. (2014). The intriguing complexities of mammalian gene regulation: How to link enhancers to regulated genes. Are we there yet? FEBS Letters, 588 (15), 2379–2391. https://doi.org/10.1016/j.febslet.2014.05.041.]. Factors that enhance or silence gene transcription also play an important role in gene expression. Both can influence the gene expression regardless of the location in the genome [Pennacchio et al. 2013Pennacchio, L.A., Bickmore, W., Dean, A., Nobrega, M.A., Bejerano, G. (2013). Enhancers: Five Essential Guestions. Nat. Rev. Gen., 14(4), 288–295. https://doi.org/10.1038/nrg3458.]. The number of transcripts in a cell that code for a particular protein is not equal to the amount of protein produced. Changes in the efficiency of protein synthesis among others depended on the elements controlling the translation, mostly located in the UTR sequences [Holcik and Pestova 2007Holcik, M., Pestova, T.V. (2007). Translation mechanism and regulation: old players, new concepts. Meeting on Translational Control and Non-Coding RNA. EMBO Rep., 8(7), 639–643. https://doi.org/10.1038/sj.embor.7400988.].
In all vertebrates, α- and β-globin genes play the same role in the body, i.e. oxygen transport in erythroid cells. However, the division of the globin gene into two families (α- and β-globin), and their location on different chromosomes caused large differences in the molecular mechanism of their expression regulation. In contrast to the family of β-globin genes, α-globins do not have characteristic promoter sequences such as the TATA box core promoter (5′-TATAAA-3′), or the inseparable CAT box regulatory sequence (5′-GCCAATCT-3′). Different cis-regulatory elements of both globin families are related to the possibility of transcription factor access. In contrast to the β-globin gene, α-globin genes are constantly exposed to transcription factors in the open chromatin structure. The absence of promoter sequences (TATA or CAT) and the location of α-globin genes near CpG islands make them similar to housekeeping genes [Forget and Hardison 2009Forget, B.G., Hardison, R.C. (2009). The normal structure ad regulation of globin gene cluster, Disorders of Hemoglobin. [In] M. Steinberg, B. Forget, D. Higgs, & D. Weatherall, Disorders of Hemoglobin: Genetics, Pathophysiology, and Clinical Management; Chapter 3, pp. 46-61. Cambridge University Press. https://doi.org/10.1017/cbo9780511596582.007.].
The coding sequence (with introns and UTRs) of the pigeon αA-globin gene is 828 base pairs. The gene consists of three exons. Non-translated regions have a length of 40 bp (5′UTR) and 100 bp (3′UTR) [NW_004973488.1]. The mRNA half-life is relatively long and ranges from 24 to 60 hours [Lodish and Small 1976Lodish, H.F., Small, B. (1976). Different lifetimes of reticulocyte messenger RNA. Cell, 7(1), 59–65. https://doi.org/10.1016/0092-8674(76)90255-5., Ross and Sullivan 1985Ross, J., Sullivan, T.D. (1985). Half-lives of beta and gamma globin messenger RNAs and of protein synthetic capacity in cultured human reticulocytes. Blood, 66(5), 1149–1154.]. The 3′UTR region plays a significant role in gene activity and post-transcriptional gene expression [Hilleren and Parker 1999Hilleren, P., Parker, R. (1999). Mechanisms of mRNA surveillance in eukaryotes. Annu. Rev. Genet., 33(1), 229–260. https://doi.org/10.1146/annurev.genet.33.1.229., Mitchell and Tollervey 2000Mitchell, P., Tollervey, D. (2000). mRNA stability in eukaryotes. Curr. Opin. Genet. Dev., 10(2), 193–198. https://doi.org/10.1016/s0959-437x(00)00063-0.], mainly by modulating mRNA stability by trans-regulatory or non-coding RNAs [Liebhaber and Russell 1998Liebhaber, S.A., Russell, J.E. (1998). Expression and developmental control of the human alpha-globin gene cluster. Ann. N. Y. Acad. Sci., 850, 54–63. https://doi.org/10.1111/j.1749-6632.1998.tb10462.x., Moss 2000Moss, E.G. (2000). Non-coding RNAs: lightning strikes twice. Curr. Biol., 10(12), 436–439. https://doi.org/10.1016/s0960-9822(00)00528-5.]. The 3′UTR region of the α-globin mRNA contains a CRE (C-rich element) stabilizing element called α-complex [Wang et al. 1995Wang, X., Kiledjian, M., Weiss, I.M., Liebhaber, S.A. (1995). Detection and characterization of a 3′ untranslated region ribonucleoprotein complex associated with human alpha-globin mRNA stability. Mol. Cell. Biol., 15(3), 1769–1777. https://doi.org/10.1128/mcb.15.3.1769., Wang and Kiledjian 2000Wang, Z., Kiledjian, M. (2000). The poly (A)-binding protein and an mRNA stability protein jointly regulate an endoribonuclease activity. Mol. Cell. Biol., 20(17), 6334–6341. https://doi.org/10.1128/mcb.20.17.6334-6341.2000.]. This complex of riboproteins correlates with the stability of mRNA by binding to the 3′UTR of the gene [Weiss and Liebhaber 1995Weiss, I.M., Liebhaber, S.A. (1995). Erythroid cell-specific mRNA stability elements in the alpha 2-globin 3′ nontranslated region. Mol. Cell. Biol., 15(5), 2457–2465. https://doi.org/10.1128/mcb.15.5.2457., Wang et al. 1999Wang, Z., Day, N., Trifillis, P., Kiledjian, M. (1999). An mRNA stability complex functions with poly(A)-binding protein to stabilize mRNA in vitro. Mol. Cell. Bio., 19(7), 4552–4560. https://doi.org/10.1128/mcb.19.7.4552.].
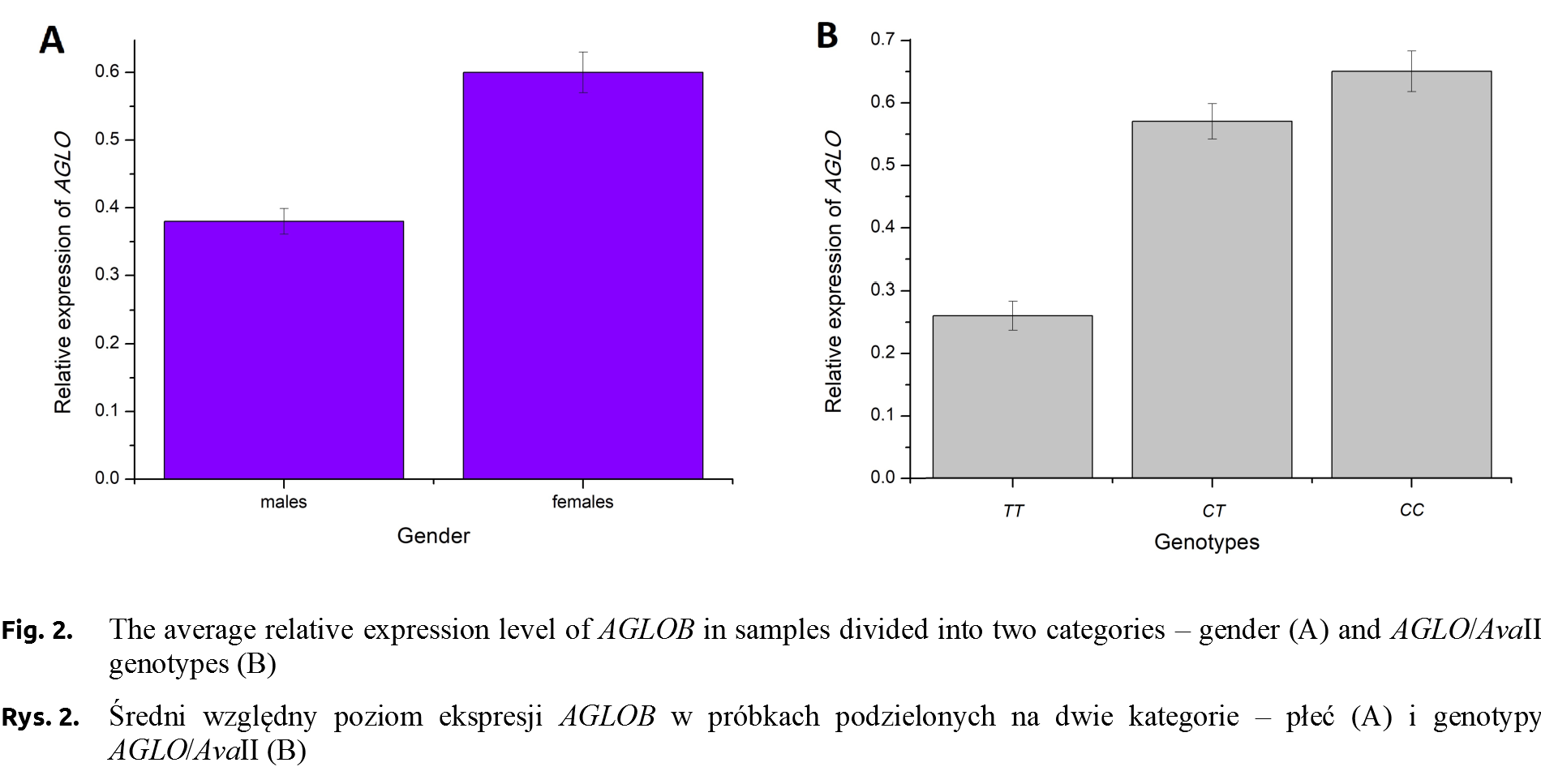
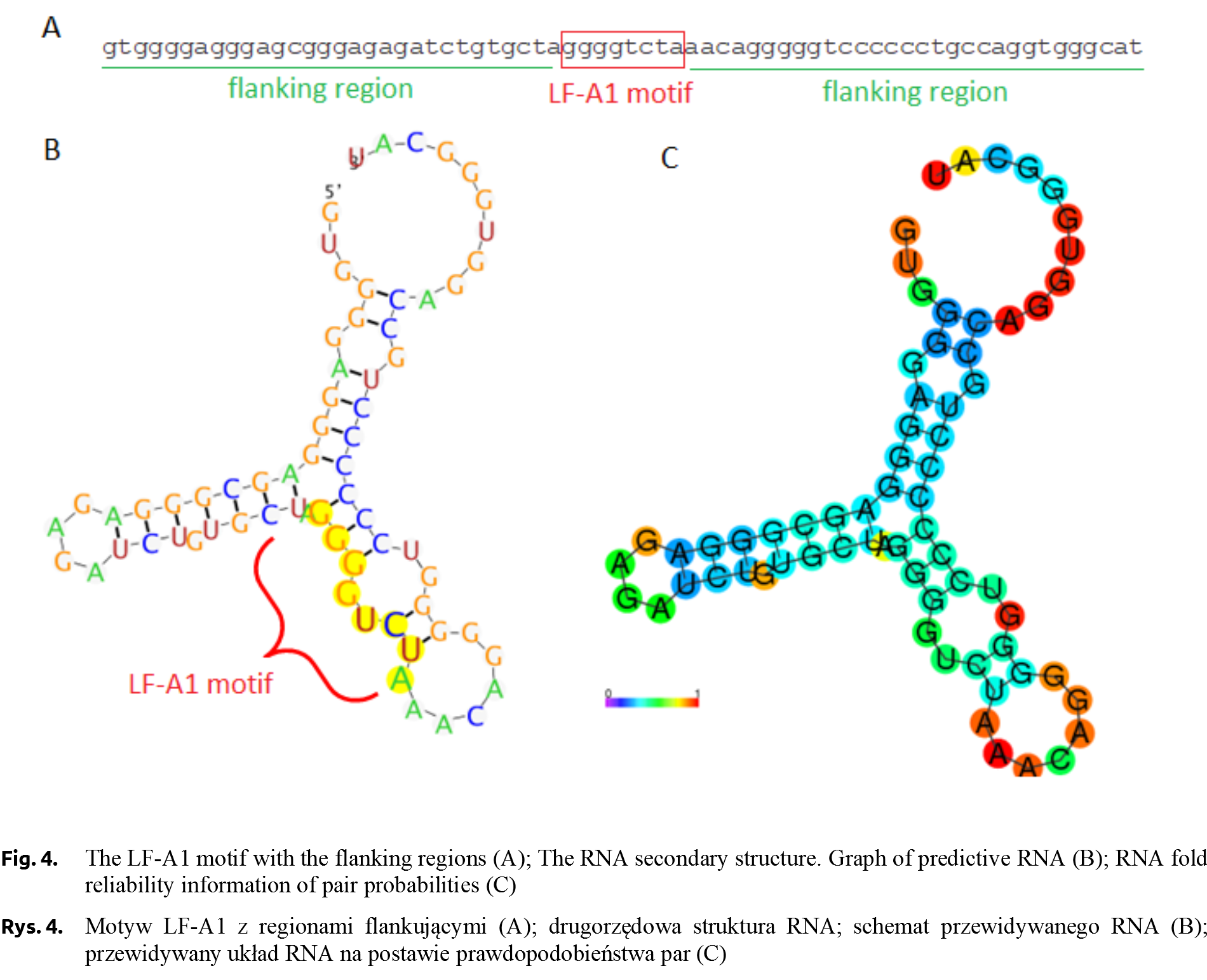
The sequences of the two homozygote genotypes (g.5768C>T) were subjected to bioinformatics analysis using the RegRNA 2.0 program [Chang et al. 2013Chang, T.H., Huang, H.Y., Hsu, J.B., Weng, S.L., Horng, J.T., Huang, H.D. (2013). An enhanced computational platform for investigating the roles of regulatory RNA and for identifying functional RNA motifs. BMC Bioinformatics, 14, Suppl. 2: S4.], in order to locate cis-regulatory regions. The analysis showed the presence of a regulatory motif for the LF-A1 transcription factor for TT genotype (Fig. 3). The occurrence of this motif was not confirmed in CC genotype. The analysis of individuals carrying different AGLOB genotypes (g.5768C>T) showed differences in the relative expression level, with the highest value was recorded for AGLOBCC ones.
The LF-A1 transcription factor, a protein with a molecular weight of about 40 kDa, binds to an 8-nucleotide regulatory motif with a TGGACT/CT/C or TGGCCC consensus sequence (Fig. 4). This fragment is identified in the promoter regions of many liver-specific genes, e.g. in the promoter sequence of haptoglobin or apolipoprotein [Ramji et al. 1991]. According to TiGER (Tissue-Specific Gene Expression and Regulation), LF-A1 factor has 30 co-regulating transcription factors, including tissue-specific ones characteristic of the brain, muscles or bone tissue [Liu et al. 2008Liu, X., Yu, X., Zack, D.J., Zhu, H., Qian, J. (2008). TiGER: a database for tissue-specific gene expression and regulation. BMC Bioinformatics, 9, 271. https://doi.org/10.1186/1471-2105-9-271.]. The LF-A1 factor occurring in TT individuals may effect on the expression of the pigeon αA-globin gene, however, there is no evidence at the current stage of research. Recently, a very low frequency of allele T (0.077) and only CT genotype detected (no TT individuals) in the group of homing pigeons was reported [Dybus et al. 2018Dybus, A., Proskura, W., Pawlina, E., Nowak, B. (2018). Associations between polymorphisms in the myostatin, αA-globin and lactate dehydrogenase B genes and racing performance in homing pigeons. Vet. Med., 63(8), 390–394. https://doi.org/10.17221/149/2017-vetmed.].
In conclusion, the effect of the SNP in the 3′ flanking region of the αA-globin gene on the level of its expression has been demonstrated. The highest expression was observed in individuals carrying the common CC genotype, while the lowest was in a very rare TT one. The fragment of the αA-globin gene under the study was analysed for the presence of cis-regulatory elements. The analysis showed the presence of a regulatory motif for the LF-A1 transcription factor in TT homozygous individuals. The occurrence of this motif has not been confirmed in individuals with the CC genotype. Lower expression level of the αA-globin gene in TT pigeons may reduce their racing performance.
Received: 10 Mar 2019
Accepted: 28 Apr 2019
Published online: 27 Jun 2019
Accesses: 1333
Jędrzejczak-Silicka, M., Dudaniec, K., Dybus, A., (2019). Association of alpha-a globin gene polymorphism with its expression level in racing pigeons. Acta Sci. Pol. Zootechnica, 18(1), 19–26. DOI: 10.21005/asp.2019.18.1.03.